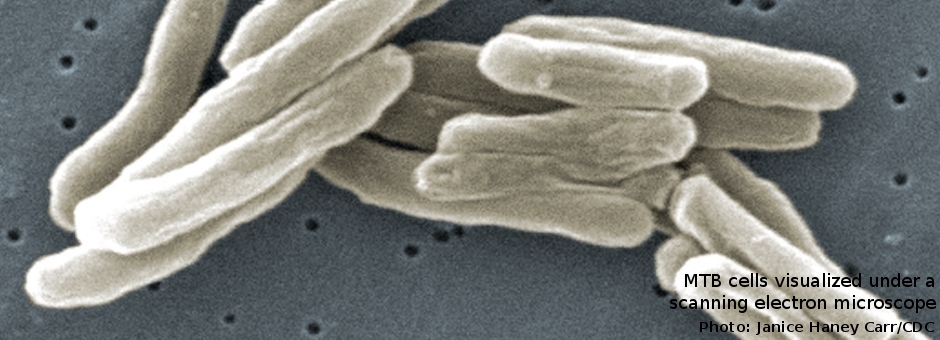
MycPermCheck is a freely accessible online tool for permeability prediction of small molecules against Mycobacterium tuberculosis. Basis of the prediction program is a logistic regression model of the physico-chemical properties of permeable substances.
In a few clicks any QikProp CSV output file may be used as an input for MycPermCheck. The file is processed by MycPermCheck and the results are visualized for easy interpretation.
Copyright © 2012 - All Rights Reserved - www.mycpermcheck.aksotriffer.pharmazie.uni-wuerzburg.de
Template by OS Templates
